GRABNES-code¶
Please check the Table of Contents on the left for quick acces to a specific module.
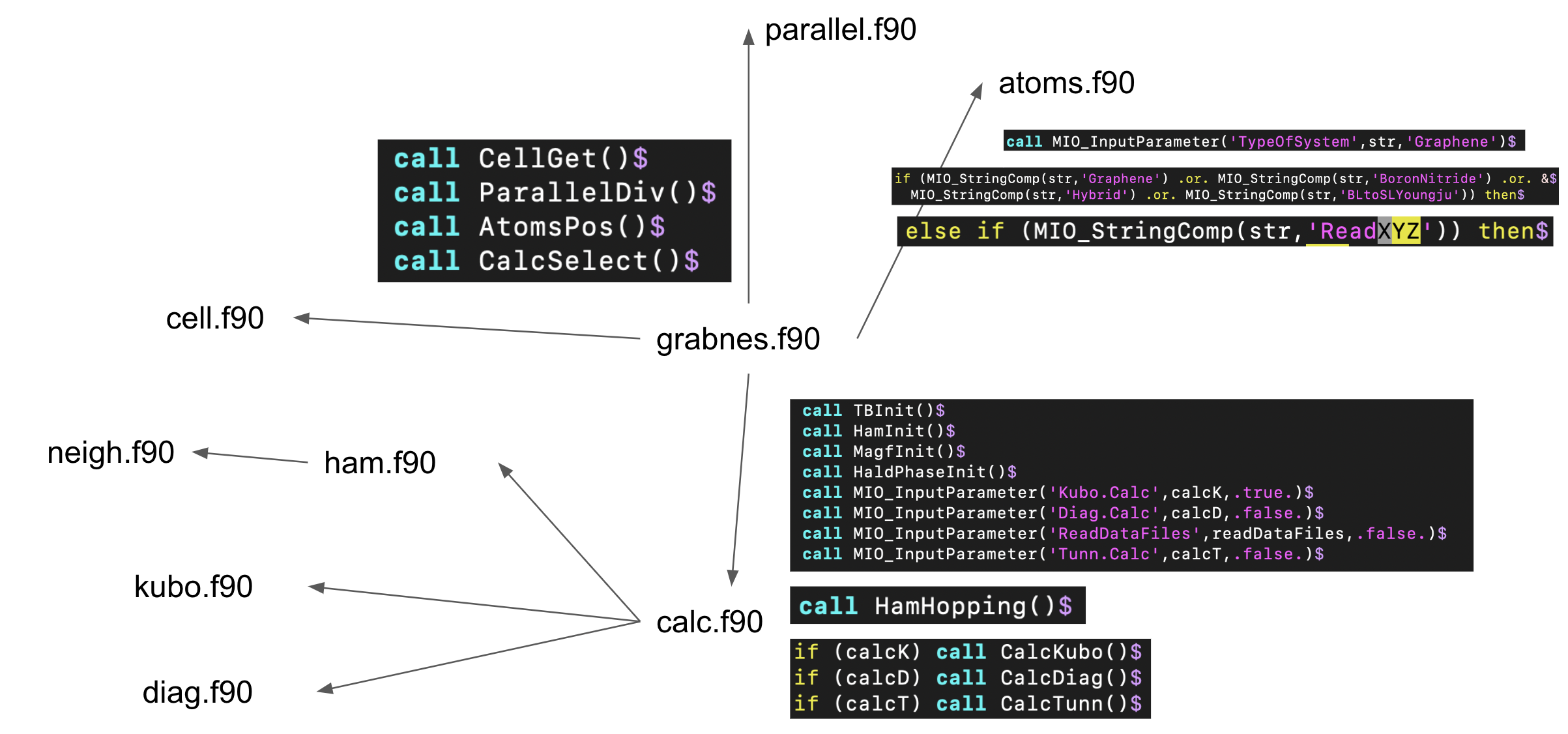
grabnes.f90¶
-
program
grabnes¶ testing sphinx
- Use
grabnes_start(grabnesstart()),grabnes_end(grabnesend()),atoms(atomspos()),cell(cellget()),calc(calcselect()),parallel(paralleldiv())- Call to
ham.f90¶
Quick access
- Variables
dcc,gpzeeman,gzeeman,h0,h0bottom,h0top,habimag,habreal,ho,hopp,hstr,indexofbigbubblecenter,moirestrain,nspin,pzterm,randomstrain,spin,strfactor,zterm- Routines
cosine_transform_data(),cosine_transform_inverse(),cross(),diago(),diago2(),diagoconj(),diagogbn(),diagoh0fromaabb(),diagohzfromaabb(),diagotbg(),distancedependentc(),fitfunsrivani(),hamhopping(),haminit(),hamonsite(),harmonicapprox(),interlayerbl(),interlayerblaa(),interlayerblab(),interlayerblba(),offdiago(),offdiago2(),offdiagogbn(),offdiagotbg(),r8vec_uniform_01(),randominrange(),timestamp()
Needed modules
mio
Variables
-
ham/dcc[real,private/save]¶
-
-
ham/gpzeeman[real,private/save]¶
-
-
ham/gzeeman[real,private/save]¶
-
-
ham/h0(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/h0bottom(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/h0top(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/habimag(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/habreal(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/ho(*) [real,pointer/public/optional/default=>null()]¶
-
-
ham/hopp(*,*) [complex,pointer/public/optional/default=>null()]¶
-
-
ham/hstr[real,private/save]¶
-
-
ham/indexofbigbubblecenter[integer,private]¶
-
-
ham/moirestrain[logical,private/save]¶
-
-
ham/nspin[integer,public/save]¶
-
-
ham/pzterm[logical,private/save]¶
-
-
ham/randomstrain[logical,private/save]¶
-
-
ham/spin[integer,private/save]¶
-
-
ham/strfactor[real,private/save]¶
-
-
ham/zterm[logical,private/save]¶
-
Subroutines and functions
-
subroutine
ham/haminit()¶ Initizalizes the Hamiltonian. It calls the neighbor subroutine and calls the __HamOnSite__ subroutine. The HamHopping routine is called at a higher level, i.e. in the calc.f90 file.
- Use
neigh(fastnnnotsquarenotrectangle()),atoms(atomssetfrac(),displacements()),interface(interfacepot2()),cell(ag(),ucell(),abn(),scell()),tbpar(g0(),tbnn()),gauss(gaussheight()),math- Called from
- Call to
fastnn(),atomssetcart(),fastnnnotsquarebulksmall(),fastnnnotsquarebulk(),fastnnnotsquaresmall(),fastnnnotsquare(),atomssetfrac(),fastnnnotsquarenotrectangle(),neighlist(),hamonsite(),gaussheight()
-
subroutine
ham/hamonsite()¶ This routine controls the onsite energy terms of the Hamiltonian. The values are stored in H0 and one must always make sure to update rather than reinitialize this value to avoid losing the additive approach when different onsite energy terms exist in the same Hamiltonian
- Use
atoms(phiforeffectivemodel(),interlayerdistances(),displacements()),tbpar(e0_c(),g0(),e0_b(),e0_n(),e0_c1(),e0_c2()),cell(scell(),ag(),abn()),constants(twopi(),pi()),parallel(ndiv(),procid()),gauss(gausspot(),gaussheight()),moireblshift(taux1(),tauy1(),taux2(),tauy2()),interface(interfacepot1()),neigh(neighd())- Called from
- Call to
atomssetfrac(),atomssetcart(),fitfunsrivani(),diagotbg(),harmonicapprox(),distancedependentc(),diagoh0fromaabb(),diago(),diagohzfromaabb(),randominrange(),gausspot()
-
subroutine
ham/randominrange(rand, rand2, low, high)¶ Rescale a random value to be inside a given range
- Parameters
rand [real,in]
rand2 [real,out]
low [real,in]
high [real,in]
- Called from
-
subroutine
ham/fitfunsrivani(p1, p2, p3, dist, cjj)¶ Obtain the interlayer distance dependent parametrization of the intralayer moire terms in tBG
- Parameters
p1 [real] :: parabolic function coefficient
p2 [real] :: parabolic function coefficient
p3 [real] :: parabolic function coefficient
dist [real] :: distance separating the two layers
cjj [real] :: fitted value
- Called from
-
subroutine
ham/distancedependentc(cd, c0, bfactor, z, z0)¶ Obtain a distance dependent reparametrization of the C0 coefficient following the Nat. Comm. expressions.
- Parameters
cd [real,out]
c0 [real,in]
bfactor [real,in]
z [real,in]
z0 [real,in]
- Called from
hamonsite(),diagoh0fromaabb(),diagohzfromaabb(),hamhopping()
-
subroutine
ham/diagoh0fromaabb(h0, dx, dy, z, moireprefactor)¶ Obtain H0 from the HAA and HBB quantities. Useful when one needs a different representation.
- Parameters
h0 [real,out]
dx [real]
dy [real]
z [real,in]
moireprefactor [real,in]
- Use
- Called from
- Call to
-
subroutine
ham/diagohzfromaabb(hz, dx, dy, z, moireprefactor)¶ Obtain Hz from the HAA and HBB quantities. Useful when one needs a different representation.
- Parameters
hz [real,out]
dx [real]
dy [real]
z [real,in]
moireprefactor [real,in]
- Use
- Called from
- Call to
-
subroutine
ham/diago(hdjj, dx, dy, cjj, phijj)¶ Obtain the diagonal terms for the GBN effective model. This works for H0 and Hz.
- Parameters
hdjj [real,out]
dx [real]
dy [real]
cjj [real,in]
phijj [real,in]
- Use
- Called from
-
subroutine
ham/diagogbn(hdjj, dx, dy, cjj, phijj, cjj0)¶ Obtain the diagonal terms for the bilayer GBN model.
-
subroutine
ham/diagoconj(hdjj, dx, dy, cjj, phijj)¶ Obtain the complex conjugate expression for the diagonal terms of the GBN effective model. Obsolete.
-
subroutine
ham/diago2(hdjj, dx, dy, cjj, phijj, cjj0)¶ Same as the diago routine, but allows to include a Cjj0 term (equal to the average of the three high symmetry stacking configurations that are used for the Harmonic approximation).
-
subroutine
ham/offdiago(hodjj, dx, dy, cjj, phijj)¶ Obtain the offdiagonal terms for the GBN effective model. This yields the HAB term that can be used to obtain the virtual strain correction to the first nearest neighbor hopping terms.
- Parameters
hodjj [complex,out]
dx [real,in]
dy [real,in]
cjj [real,in]
phijj [real,in]
- Use
- Called from
-
subroutine
ham/offdiago2(hodjj, dx, dy, cjj, phijj)¶ - Parameters
hodjj [complex,out]
dx [real,in]
dy [real,in]
cjj [real,in]
phijj [real,in]
- Use
- Called from
-
subroutine
ham/offdiagogbn(hodjj, dx, dy, cjj, phijj)¶ - Parameters
hodjj [complex,out]
dx [real,in]
dy [real,in]
cjj [real,in]
phijj [real,in]
- Use
- Called from
-
subroutine
ham/offdiagotbg(hodjj, dx, dy, cjj, phijj)¶ - Parameters
hodjj [complex,out]
dx [real,in]
dy [real,in]
cjj [real,in]
phijj [real,in]
- Use
- Called from
-
subroutine
ham/diagotbg(hodjj, dx, dy, cjj, phijj, cjj0)¶ - Parameters
hodjj [real,out]
dx [real,in]
dy [real,in]
cjj [real,in]
phijj [real,in]
cjj0 [real,in]
- Use
- Called from
-
subroutine
ham/harmonicapprox(dx, dy, a, b, c, hjj)¶ - Parameters
dx [real,in]
dy [real,in]
a [real,in]
b [real,in]
c [real,in]
hjj [real,out]
- Use
- Called from
-
subroutine
ham/interlayerbl(hbl, dx, dy, tab)¶
-
subroutine
ham/interlayerblab(hab, dx, dy, tbt, posorneg)¶ - Parameters
hab [complex,out]
dx [real,in]
dy [real,in]
tbt [real,in]
posorneg [real,in]
- Use
- Called from
-
subroutine
ham/interlayerblba(hba, dx, dy, tbt, posorneg)¶ - Parameters
hba [complex,out]
dx [real,in]
dy [real,in]
tbt [real,in]
posorneg [real,in]
- Use
- Called from
-
subroutine
ham/interlayerblaa(haa, dx, dy, tbt, posorneg)¶ - Parameters
haa [complex,out]
dx [real,in]
dy [real,in]
tbt [real,in]
posorneg [real,in]
- Use
- Called from
-
subroutine
ham/hamhopping()¶ - Use
neigh(nneigh(),neighd(),nlist(),neighcell(),nradii()),atoms(species(),dintlay(),rat(),frac(),atomssetcart(),atomssetfrac(),inode1(),inode2()),tbpar(gintlay(),gn(),tbnn(),g0()),magf(magfield(),bmag(),haldanephase()),constants(pi(),fluxq(),cmplx_i(),twopi(),ub()),cell(ag(),abn()),gauss(gausspotdefinedpositions()),moireblshift(taux1(),tauy1(),taux2(),tauy2()),math,name(prefix(),sysname())- Called from
- Call to
atomssetfrac(),atomssetcart(),distancedependentc(),offdiago(),offdiago2(),fitfunsrivani(),offdiagotbg(),offdiagogbn(),interlayerblaa(),interlayerblab(),interlayerblba(),cross(),gausspotdefinedpositions()
-
subroutine
ham/cosine_transform_data(n, d, c)¶ - Options
n [integer,optional/default=len(d)]
- Parameters
d (n) [real]
c (n) [real]
- Call to
-
subroutine
ham/cosine_transform_inverse(n, c, d)¶ - Options
n [integer,optional/default=len(c)]
- Parameters
c (n) [real]
d (n) [real]
- Call to
-
subroutine
ham/r8vec_uniform_01(n, seed, r)¶ - Options
n [integer,optional/default=len(r)]
- Parameters
seed [integer]
r (n) [real]
- Call to
-
function
ham/cross(a, b)¶ - Parameters
a (3) [real,in]
b (3) [real,in]
- Return
axb (3) [real]
- Called from
hamhopping(),cosine_transform_data(),cosine_transform_inverse(),r8vec_uniform_01(),timestamp()
atoms.f90¶
Quick access
- Variables
dintlay,displacements,frac,in1,in2,indxdiv,indxnode,inode1,inode2,interlayerdistances,layerindex,nat,natbn,natc,natc1,natc2,natc3,nel,phiforeffectivemodel,procat,rat,ratbn,ratc,ratc1,ratc2,ratc3,ratinit,rattemp,secat,species,speciestemp- Routines
atomsconstruct(),atomsconstructasymm(),atomsconstructbasedonfastnn(),atomsorder(),atomspart(),atomspos(),atomsribbons(),atomsrotate(),atomssetcart(),atomssetcarttemp(),atomssetfrac(),atomssetfractemp(),atomssort(),gcd()
Needed modules
miomath
Variables
-
atoms/dintlay[real,public]¶
-
-
atoms/displacements(*,*) [real,pointer/public]¶
-
-
atoms/frac[logical,save/public/optional/default=.true.]¶
-
-
atoms/in1[integer,save/public]¶
-
-
atoms/in2[integer,save/public]¶
-
-
atoms/indxdiv(*) [integer,pointer/public]¶
-
-
atoms/indxnode(*) [integer,pointer/public]¶
-
-
atoms/inode1[integer,save/public]¶
-
-
atoms/inode2[integer,save/public]¶
-
-
atoms/interlayerdistances(*) [real,pointer/public]¶
-
-
atoms/layerindex(*) [integer,pointer/public]¶
-
-
atoms/nat[integer,save/public]¶
-
-
atoms/natbn[integer,save/public]¶
-
-
atoms/natc[integer,save/public]¶
-
-
atoms/natc1[integer,save/public]¶
-
-
atoms/natc2[integer,save/public]¶
-
-
atoms/natc3[integer,save/public]¶
-
-
atoms/nel[real,public]¶
-
-
atoms/phiforeffectivemodel[real,public]¶
-
-
atoms/procat(*) [integer,pointer/public]¶
-
-
atoms/rat(*,*) [real,pointer/public]¶
-
-
atoms/ratbn(*,*) [real,pointer/public]¶
-
-
atoms/ratc(*,*) [real,pointer/public]¶
-
-
atoms/ratc1(*,*) [real,pointer/public]¶
-
-
atoms/ratc2(*,*) [real,pointer/public]¶
-
-
atoms/ratc3(*,*) [real,pointer/public]¶
-
-
atoms/ratinit(*,*) [real,pointer/public]¶
-
-
atoms/rattemp(*,*) [real,pointer/public]¶
-
-
atoms/secat(*) [integer,pointer/public]¶
-
-
atoms/species(*) [integer,pointer/public]¶
-
-
atoms/speciestemp(*) [integer,pointer/public]¶
-
Subroutines and functions
-
subroutine
atoms/atomspos()¶ - Use
math,parallel(ndiv(),nth(),procid()),cell(scell(),scell2(),ag(),ucell(),rcell(),volume(),area(),abn()),constants(pi(),twopi()),hybrid(hybridgen()),name(prefix())- Called from
- Call to
atomsconstruct(),atomsconstructasymm(),atomssetcart(),atomssetfrac(),atomsribbons(),atomssetfractemp(),atomsorder(),hybridgen()
-
subroutine
atoms/atomsconstruct(x, mm, angle, label, d, yshift)¶ - Parameters
x (,) [real,out,pointer]
mm (2) [integer,in]
angle [real,in]
label [character,in]
d [real,in]
yshift [real,in]
- Use
- Called from
- Call to
-
subroutine
atoms/atomsconstructbasedonfastnn(x, mm, angle, label, d, yshift, natoms)¶
-
subroutine
atoms/atomsconstructasymm(x, mm, mm2, angle, label, d, yshift)¶ - Parameters
x (,) [real,out,pointer]
mm (2) [integer,in]
mm2 (2) [integer,in] :: I think this takes care of y direction (second column) of m matrix
angle [real,in]
label [character,in]
d [real,in]
yshift [real,in]
- Use
- Called from
- Call to
-
subroutine
atoms/atomsribbons(type_bn, nc, nbn, n, x, sp)¶ - Parameters
type_bn [character,in]
nc [integer,in]
nbn [integer,in]
n [integer,in]
x (3,n) [real,out]
sp (n) [integer,out]
- Called from
-
subroutine
atoms/atomssetcart()¶ - Use
- Called from
atomspos(),calcselect(),calckubo(),calctunn(),diagspectralweightweikuinequivalentold(),diagspectralweightweikuinequivalent(),diagspectralweightweikuinequivalentlee(),diagspectralweightweikuinequivalentnishi(),gausspot(),gausspotdefinedpositions(),gaussheight(),haminit(),hamonsite(),hamhopping(),interfacepot2(),neighlist(),neighlistold(),fastnnnotsquarenotrectangle()
-
subroutine
atoms/atomssetfrac()¶ - Use
- Called from
atomspos(),calckubo(),diagspectralweightweiku(),haminit(),hamonsite(),hamhopping()
-
subroutine
atoms/atomssetfractemp()¶ - Use
- Called from
-
subroutine
atoms/atomsorder()¶
-
subroutine
atoms/atomssort(array1, array2, array3, array4)¶ - Parameters
array1 (*) [integer,inout]
array2 (*) [integer,inout]
array3 (,) [real,inout]
array4 (*) [integer,inout]
- Called from
-
subroutine
atoms/atomspart(array1, array2, array3, array4, mrk)¶ - Parameters
array1 (*) [integer,inout]
array2 (*) [integer,inout]
array3 (,) [real,inout]
array4 (*) [integer,inout]
mrk [integer,out]
-
function
atoms/gcd(i1, i2)¶ - Parameters
i1 [integer,in]
i2 [integer,in]
- Return
gcd [integer]
- Called from
atomsconstruct(),atomsconstructbasedonfastnn(),atomsconstructasymm()
calc.f90¶
Quick access
- Routines
Needed modules
mio
Variables
Subroutines and functions
-
subroutine
calc/calcselect()¶ - Use
ham(haminit(),hamhopping(),hamonsite(),hopp()),tbpar(tbinit()),magf(magfinit(),mbi(),mbf(),magfvalue(),mstep(),haldphaseinit(),mphii(),mphif(),haldphasevalue(),mphistep(),magfield(),bmag()),moireblshift(msi(),msf(),msstep(),moireblshiftinit(),moireblshiftvalue()),atoms(nat(),rat(),frac(),atomssetcart()),neigh(nneigh(),neighcell(),nlist(),maxneigh()),constants,cell(ucell()),math- Called from
- Call to
tbinit(),haminit(),hamonsite(),magfinit(),magfvalue(),hamhopping(),calckubo(),calcdiag(),haldphaseinit(),haldphasevalue(),atomssetcart(),calctunn()
-
subroutine
calc/calckubo()¶ - Use
atoms(atomssetfrac(),atomssetcart(),species()),kuboarrays,kubo(kuboinitwf(),kuboinitwfpdos(),kubodos(),kubotevol(),kuboinitwflayerdos()),kubosubs(kubointerval(),kubocn()),neigh(rcvlist(),sndsz()),ham(h0(),hopp()),cell(scell(),ag()),tbpar(g0()),name(prefix(),sysname())- Called from
- Call to
atomssetcart(),kuboinitwflayerdos(),kubodos(),kuboinitwfpdos(),atomssetfrac(),kuboinitwf(),kubointerval(),kubocn(),kubotevol()
-
subroutine
calc/calctunn()¶
-
subroutine
calc/calcdiag()¶ - Use
diag(diaginit(),diagdos(),diagpdos(),diagbands(),diag3dbands(),diagbandsg(),diagbandsaroundk(),diagspectralfunction(),diagspectralfunctionkgrid(),diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagbandsrashba()),atoms(nat()),ham(nspin()),scf(scfgetcharge())- Called from
- Call to
diaginit(),scfgetcharge(),diagpdos(),diagdos(),diagbandsg(),diagbandsaroundk(),diagbandsrashba(),diagbands(),diag3dbands(),diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut()
cell.f90¶
Quick access
Needed modules
mio
Variables
-
cell/abn[real,save/public]¶
-
-
cell/ag[real,save/public]¶
-
-
cell/area[real,save/public]¶
-
-
cell/rcell(3,3) [real,save/public]¶
-
-
cell/scell[integer,save/public]¶
-
-
cell/scell2[integer,save/public]¶
-
-
cell/ucell(3,3) [real,save/public]¶
-
-
cell/volume[real,save/public]¶
-
Subroutines and functions
constants.f90¶
Quick access
Needed modules
mio
Variables
-
constants/cmplx_0[complex,public/parameter/optional/default=(0.0_dp,0.0_dp)]¶
-
-
constants/cmplx_1[complex,public/parameter/optional/default=(1.0_dp,0.0_dp)]¶
-
-
constants/cmplx_i[complex,public/parameter/optional/default=(0.0_dp,1.0_dp)]¶
-
-
constants/fluxq[real,public/parameter/optional/default=hplanck/qe]¶
-
-
constants/hbar[real,public/parameter/optional/default=hplanck/twopi]¶
-
-
constants/hplanck[real,public/parameter/optional/default=6.62606896d-34]¶
-
-
constants/pi[real,public/parameter/optional/default=3.14159265358979323846264338327950288419716939937510582097494459230781640_dp]¶
-
-
constants/qe[real,public/parameter/optional/default=1.602176565d-19]¶
-
-
constants/twopi[real,public/parameter/optional/default=2.0_dp*pi]¶
-
-
constants/ub[real,public/parameter/optional/default=5.7883818066d-5]¶
-
diag.f90¶
Quick access
- Variables
- Routines
convolve(),diag3dbands(),diagbands(),diagbandsaroundk(),diagbandsg(),diagbandsrashba(),diagdos(),diagham(),diaghamarpack(),diaghampdos(),diaghamrashba(),diaghamwf(),diaginit(),diagpdos(),diagspectralfunction(),diagspectralfunctionkgrid(),diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagspectralfunctionkgridinequivalentenergycutnickdale(),diagspectralweightnishi(),diagspectralweightweiku(),diagspectralweightweikuinequivalent(),diagspectralweightweikuinequivalentlee(),diagspectralweightweikuinequivalentnishi(),diagspectralweightweikuinequivalentold(),matinv3()
Needed modules
mio
Variables
-
diag/dwork(*) [real,private/pointer/optional/default=>null()]¶
-
-
diag/efermi[real,private/save/optional/default=0.0_dp]¶
-
-
diag/emax[real,private/save/optional/default=20.0_dp]¶
-
-
diag/emin[real,private/save/optional/default=-50.0_dp]¶
-
-
diag/lwork[integer,private]¶
-
-
diag/zwork(*) [complex,private/pointer/optional/default=>null()]¶
-
Subroutines and functions
-
subroutine
diag/diaginit(n)¶ - Parameters
n [integer,in]
- Called from
-
subroutine
diag/diagdos()¶
-
subroutine
diag/diagpdos()¶
-
subroutine
diag/diag3dbands()¶
-
subroutine
diag/diagbands()¶
-
subroutine
diag/diagbandsrashba()¶
-
subroutine
diag/diagbandsaroundk()¶
-
subroutine
diag/diagbandsg()¶
-
subroutine
diag/diagspectralfunction()¶
-
subroutine
diag/diagspectralfunctionkgrid()¶
-
subroutine
diag/diagspectralfunctionkgridinequivalent()¶ - Use
cell(rcell(),ucell()),atoms(nat(),frac(),layerindex()),ham(h0(),hopp(),nspin()),neigh(nlist(),nneigh(),neighcell(),maxneigh()),name(prefix()),tbpar(g0()),constants(pi(),twopi()),math- Called from
- Call to
diagspectralweightweikuinequivalentold(),diagspectralweightweikuinequivalent(),diagspectralweightweikuinequivalentlee(),diagspectralweightweikuinequivalentnishi(),convolve()
-
subroutine
diag/diagspectralfunctionkgridinequivalentenergycut()¶ - Use
cell(rcell(),ucell()),atoms(nat(),frac(),layerindex()),ham(h0(),hopp(),nspin()),neigh(nlist(),nneigh(),neighcell(),maxneigh()),name(prefix()),tbpar(g0()),constants(pi(),twopi()),math- Called from
- Call to
diagspectralweightweikuinequivalentold(),diagspectralweightweikuinequivalent(),diagspectralweightweikuinequivalentnishi(),convolve()
-
subroutine
diag/diagspectralfunctionkgridinequivalentenergycutnickdale()¶ - Use
cell(rcell(),ucell()),atoms(nat(),frac(),layerindex()),ham(h0(),hopp(),nspin()),neigh(nlist(),nneigh(),neighcell(),maxneigh()),name(prefix()),tbpar(g0()),constants(pi(),twopi()),math- Call to
diagspectralweightweikuinequivalentold(),diagspectralweightweikuinequivalent(),diagspectralweightweikuinequivalentnishi(),convolve()
-
subroutine
diag/diagham(n, ns, is, hloc, eloc, kloc, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
hloc (n,n) [complex,out]
eloc (n) [real,out]
kloc (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species()),tbpar(u())- Called from
diagdos(),diag3dbands(),diagbands(),diagbandsaroundk(),diagbandsg()
-
subroutine
diag/diaghamwf(n, ns, is, hloc, eloc, kloc, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
hloc (n,n) [complex,out]
eloc (n) [real,out]
kloc (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species()),tbpar(u())- Called from
-
subroutine
diag/diaghampdos(n, ns, is, hloc, eloc, kloc, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
hloc (n,n) [complex,out]
eloc (n) [real,out]
kloc (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species()),tbpar(u())- Called from
-
subroutine
diag/diaghamrashba(n, ns, is, hloc, eloc, kloc, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
hloc (2 * n,2 * n) [complex,out] :: double the output Hamiltonian size to account for spin
eloc (2 * n) [real,out] :: same for eigenergies
kloc (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in] :: Repeat initial H0 a second time
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),layerindex()),tbpar(u()),neigh(neighd())- Called from
-
subroutine
diag/diaghamarpack(n, ns, is, hloc, eloc, kloc, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
hloc (n,n) [complex,out]
eloc (n) [real,out]
kloc (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species()),tbpar(u())
-
subroutine
diag/diagspectralweightnishi(n, ns, is, pkc, e, k, kg, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
pkc (n) [complex,out]
e (n) [real,out]
k (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac()),tbpar(u()),neigh(maxneigh())
-
subroutine
diag/diagspectralweightweiku(n, ns, is, pkcloc, e, k, kg, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)] :: NOT the sum over eigenvectors. Pick one eigenvector and then sum over its coefficients. Each coefficient corresponds to one orbital in Wannier (or TB) basis.
- Parameters
ns [integer,in]
is [integer,in]
pkcloc (n,2) [complex,out]
e (n) [real,out]
k (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac()),tbpar(u()),neigh(maxneigh())- Called from
- Call to
-
subroutine
diag/diagspectralweightweikuinequivalentold(n, ns, is, pkcloca, pkclocb, eloc, kptsloc, kg, cell, gcell, h0, maxn, hopp, nlist, nneigh, neighcell, topbottomratio)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
pkcloca (n,3) [complex,out]
pkclocb (n,3) [complex,out]
eloc (n) [real,out]
kptsloc (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in]
gcell (3,3) [real] :: Lattice vectors of PC (either top or bottom layer)
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
topbottomratio [real]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac(),layerindex()),tbpar(u()),neigh(maxneigh())- Called from
diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagspectralfunctionkgridinequivalentenergycutnickdale()- Call to
-
subroutine
diag/diagspectralweightweikuinequivalent(n, ns, is, pkcloca, pkclocb, eloc, kptsloc, kg, cell, gcell, h0, maxn, hopp, nlist, nneigh, neighcell, topbottomratio)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
pkcloca (n,3) [complex,out]
pkclocb (n,3) [complex,out]
eloc (n) [real,out]
kptsloc (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in]
gcell (3,3) [real] :: Lattice vectors of PC (either top or bottom layer)
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
topbottomratio [real]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac(),layerindex()),tbpar(u()),neigh(maxneigh())- Called from
diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagspectralfunctionkgridinequivalentenergycutnickdale()- Call to
-
subroutine
diag/diagspectralweightweikuinequivalentlee(n, ns, is, pkcloc1, pkcloc2, eloc, kptsloc, kg, cell, gcell, h0, maxn, hopp, nlist, nneigh, neighcell, topbottomratio)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
ns [integer,in]
is [integer,in]
pkcloc1 (n,3) [complex,out]
pkcloc2 (n,3) [complex,out]
eloc (n) [real,out]
kptsloc (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in] :: Lattice vectors of PC (either top or bottom layer)
gcell (3,3) [real]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
topbottomratio [real]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac(),layerindex()),tbpar(u()),neigh(maxneigh()),math- Called from
- Call to
-
subroutine
diag/diagspectralweightweikuinequivalentnishi(n, ns, is, pkcloc, eloc1, eloc2, kptsloc, kg, cell, gcell1, gcell2, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)] :: NOT the sum over eigenvectors. Pick one eigenvector and then sum over its coefficients. Each coefficient corresponds to one orbital in Wannier (or TB) basis.
- Parameters
ns [integer,in]
is [integer,in]
pkcloc (n,2) [complex,out]
eloc1 (n) [real,out]
eloc2 (n) [real,out]
kptsloc (3) [real,in]
kg (3) [real,in]
cell (3,3) [real,in]
gcell1 (3,3) [real]
gcell2 (3,3) [real]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
constants(cmplx_i()),interface(edgehopp(),nedgen(),edgeh(),nq(),edgeindx(),nei(),nedgecell()),scf(charge(),zch()),atoms(species(),rat(),atomssetcart(),atomssetfrac(),frac(),layerindex()),tbpar(u()),neigh(maxneigh())- Called from
diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagspectralfunctionkgridinequivalentenergycutnickdale()- Call to
-
function
diag/convolve(x, h, epts)¶ - Parameters
x (epts) [real]
h (epts) [real]
- Options
epts [integer,optional/default=len(x)]
- Return
convolve (*) [real,allocatable]
- Called from
diagspectralfunction(),diagspectralfunctionkgrid(),diagspectralfunctionkgridinequivalent(),diagspectralfunctionkgridinequivalentenergycut(),diagspectralfunctionkgridinequivalentenergycutnickdale(),convolve()- Call to
-
function
diag/matinv3(a)¶ - Parameters
a (3,3) [real,in]
- Return
b (3,3) [real]
gauss.f90¶
Quick access
- Routines
Needed modules
mio
Variables
Subroutines and functions
-
subroutine
gauss/gausspot(h)¶
interface.f90¶
Quick access
- Variables
ampb,ampeh,ampn,dampeh,edgeh,edgehopp,edgeindx,lambda,maxedgeneigh,ncellint,nedgecell,nedgen,nei,nq,rmaxint- Routines
Needed modules
mio
Variables
-
interface/ampb(4) [real,private/save]¶
-
-
interface/ampeh[real,private/save]¶
-
-
interface/ampn(4) [real,private/save]¶
-
-
interface/dampeh[real,private/save]¶
-
-
interface/edgeh(*,*) [real,pointer/public]¶
-
-
interface/edgehopp[logical,save/public]¶
-
-
interface/edgeindx(*) [integer,pointer/public]¶
-
-
interface/lambda(2,4) [real,private/save]¶
-
-
interface/maxedgeneigh[integer,private/parameter/optional/default=30]¶
-
-
interface/ncellint(2) [integer,private/save]¶
-
-
interface/nedgecell(*,*,*) [integer,pointer/public]¶
-
-
interface/nedgen(*) [integer,pointer/public]¶
-
-
interface/nei(*,*) [integer,pointer/public]¶
-
-
interface/nq[integer,save/public]¶
-
-
interface/rmaxint[real,private/save]¶
-
Subroutines and functions
kubo.f90¶
Quick access
- Routines
kubodos(),kuboinitwf(),kuboinitwflayerdos(),kuboinitwfpdos(),kubotevol()
Needed modules
Variables
Subroutines and functions
-
subroutine
kubo/kuboinitwf(psi)¶
-
subroutine
kubo/kuboinitwfpdos(psi, pdosatomnumber)¶
-
subroutine
kubo/kuboinitwflayerdos(psi, layernumber, numberoflayers, numberofatomsinlayer)¶
-
subroutine
kubo/kubodos(psi, psin, psinm1, a, b, h, h0, hopp, nlist, nrecurs, eps, nen, emin, emax)¶ - Parameters
psi (-(inode1)+1) [complex,out]
psin (-(inode1)+1) [complex,out]
psinm1 (-(inode1)+1) [complex,out]
a (nrecurs) [real,out]
b (nrecurs) [real,out]
h (-(inode1)+1) [complex,out]
h0 (-(inode1)+1) [real,in]
hopp (*,-(inode1)+1) [complex,in] ::
nlist (,-(inode1)+1) [integer,in]
nrecurs [integer,in]
eps [real,in]
nen [integer,in]
emin [real]
emax [real]
- Use
name(prefix()),kubosubs(kuborecursion(),kubofrac(),kubofermi())- Called from
- Call to
-
subroutine
kubo/kubotevol(psi, zupsi, psin, psinm1, xpnpsi, xpnpsim1, c, dt, nt, az, bz, h, h0, hopp, nlist, neighd, ac, bc, nen, emin, emax, eps, nrecurs, npol, nwr)¶ - Parameters
psi (-(inode1)+1) [complex,inout]
zupsi (-(inode1)+1) [complex,inout]
psin (-(inode1)+1) [complex,inout]
psinm1 (-(inode1)+1) [complex,inout]
xpnpsi (-(inode1)+1) [complex,inout]
xpnpsim1 (-(inode1)+1) [complex,inout]
c (npol) [complex,in]
dt [real,in]
nt [integer,in]
az (nrecurs) [real,inout]
bz (nrecurs) [real,inout]
h (-(inode1)+1) [complex,out]
h0 (-(inode1)+1) [real,in]
hopp (*,-(inode1)+1) [complex,in] ::
nlist (,-(inode1)+1) [integer,in]
neighd (,,-(inode1)+1) [real,in]
ac [real,in]
bc [real,in]
nen [integer,in]
emin [real,in]
emax [real,in]
eps [real,in]
nwr [integer,in]
- Options
nrecurs [integer,in,optional/default=len(az)]
npol [integer,in,optional/default=len(c)]
- Use
constants(cmplx_0()),name(prefix()),kubosubs(kuborecursion(),kubofrac(),kuboevol())- Called from
- Call to
kuboarrays.f90¶
Quick access
Needed modules
mio
Variables
-
kuboarrays/a(*) [real,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/b(*) [real,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/c(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/h(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/psi(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/psin(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/psinm1(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/tempd(*) [real,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/tempz(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/xpnpsi(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/xpnpsim1(*) [complex,pointer/public/optional/default=>null()]¶
-
-
kuboarrays/zupsi(*) [complex,pointer/public/optional/default=>null()]¶
-
kubosubs.f90¶
Quick access
- Routines
kubocn(),kuboevol(),kubofermi(),kubofrac(),kubointerval(),kuborecursion(),kuboupdate_d(),kuboupdate_z(),tql2()
Needed modules
Variables
Subroutines and functions
-
subroutine
kubosubs/kuborecursion(psi, psin, psinm1, nrecurs, a, b, h, h0, hopp, nlist, wr[, name])¶ - Parameters
psi (-(inode1)+1) [complex,inout]
psin (-(inode1)+1) [complex,inout]
psinm1 (-(inode1)+1) [complex,inout]
nrecurs [integer,in]
a (nrecurs) [real,out]
b (nrecurs) [real,out]
h (-(inode1)+1) [complex,out]
h0 (-(inode1)+1) [real,in]
hopp (*,-(inode1)+1) [complex,in] ::
nlist (,-(inode1)+1) [integer,in]
wr [logical,in]
name [character,in,]
- Use
kuboarrays(tempz()),constants(cmplx_0()),neigh(maxneigh(),nneigh())- Called from
-
subroutine
kubosubs/kubofrac(a, b, nrecurs, norm, eps, nen, emin, emax, name, name2)¶ - Parameters
a (nrecurs) [real,in]
b (nrecurs) [real,in]
norm [real,in]
eps [real,in]
nen [integer,in]
emin [real,in]
emax [real,in]
name [character,in]
name2 [character,in]
- Options
nrecurs [integer,in,optional/default=len(a)]
- Use
- Called from
-
subroutine
kubosubs/kubointerval(nrecurs, a, b, ac, bc)¶ - Options
nrecurs [integer,in,optional/default=len(a)]
- Parameters
a (nrecurs) [real,in]
b (nrecurs) [real,in]
ac [real,out]
bc [real,out]
- Called from
-
subroutine
kubosubs/kubocn(dt, ac, bc, npol, c)¶
-
subroutine
kubosubs/kuboevol(psi, zupsi, psin, psinm1, xpnpsi, xpnpsim1, c, h0, hopp, nlist, neighd, ac, bc, npol, prnt)¶ - Parameters
psi (-(inode1)+1) [complex,inout]
zupsi (-(inode1)+1) [complex,inout]
psin (-(inode1)+1) [complex,inout]
psinm1 (-(inode1)+1) [complex,inout]
xpnpsi (-(inode1)+1) [complex,inout]
xpnpsim1 (-(inode1)+1) [complex,inout]
c (npol) [complex,in]
h0 (-(inode1)+1) [real,in]
hopp (*,-(inode1)+1) [complex,in] ::
nlist (,-(inode1)+1) [integer,in]
neighd (,,-(inode1)+1) [real,in]
ac [real,in]
bc [real,in]
prnt [logical,in]
- Options
npol [integer,in,optional/default=len(c)]
- Use
constants(cmplx_0()),kuboarrays(tempz()),neigh(maxneigh(),nneigh())- Called from
-
subroutine
kubosubs/kuboupdate_d(array, temp)¶
-
subroutine
kubosubs/kuboupdate_z(array, temp)¶
-
subroutine
kubosubs/tql2(nm, n, d, e, z, ier)¶ QL METHOD TO DETERMINE THE EIGENVALUES AND EIGENVECTORS OF:
A SYMMETRIC TRIDIAGONAL MATRIX.
A FULL SYMMETRIC MATRIX AFTER A PREVIOUS CALL TO TRED2.
- CALLING MODE:
CALL TQL2(NM,N,D,E,Z,IER)
INPUTSS: NM (I4) 1ST DIMENSION OF MATRICES A AND Z IN CALLING PROGRAM N (I4) SIZE OF Z D (R*8) MAIN DIAGONAL (N) OF THE TRIDIAGONAL MATRIX E (R*8) SUB-DIAGONAL (N) OF THE TRIDIAGONAL MATRIX Z (R*8) TABLE (NM,N) STORING THE UNITY MATRIX IF THE TRIDIAGONAL
MATRIX IS DEFINED BY D AND E, CASE #1. FOR CASE #2, IT CONTAINS THE ELEMENTS OF THE TRANSFORMATION MATRIX AFTER A CALL TO TRED2.
OUTPUTS: D (R*8) EIGENVALUES Z (R*8) EIGENVECTORS IER (I4) ERROR CODE = 0, CONVERGENCE OK.
= L, NO CONVERGENCE FOR THE Lth EIGENVALUE
REFERENCE: J.H.WILKINSON,-C.REINSCH,R.S.MARTIN HANDBOOK FOR AUTOMATIC COMPUTATION, VOL.2, LINEAR ALGEBRA SPRINGER-VERLAG 1971.
- Parameters
nm [integer]
d (n) [real]
e (n) [real]
z (nm,n) [real]
ier [integer]
- Options
n [integer,optional/default=len(d)]
- Called from
magf.f90¶
Quick access
Needed modules
mio
Variables
-
magf/bmag[real,public/save/optional/default=0.0_dp]¶
-
-
magf/haldanephase[real,public/save]¶
-
-
magf/haldphase[logical,public/save/optional/default=.false.]¶
-
-
magf/magfield[logical,public/save/optional/default=.false.]¶
-
-
magf/mbf[integer,public/save/optional/default=0]¶
-
-
magf/mbi[integer,public/save/optional/default=0]¶
-
-
magf/mphif[integer,public/save/optional/default=0]¶
-
-
magf/mphii[integer,public/save/optional/default=0]¶
-
-
magf/mphistep[integer,public/save/optional/default=0]¶
-
-
magf/mstep[integer,public/save/optional/default=1]¶
-
Subroutines and functions
-
subroutine
magf/magfvalue(mb)¶
neigh.f90¶
Quick access
- Variables
maxneigh,neighcell,neighd,nlist,nlist2,nneigh,nodesndrcv,nradii,rcvindx,rcvlist,rcvsz,sndindx,sndlist,sndsz- Routines
fastnn(),fastnnnotsquare(),fastnnnotsquarebulk(),fastnnnotsquarebulksmall(),fastnnnotsquarenotrectangle(),fastnnnotsquaresmall(),neighindxcell(),neighlist(),neighlistold()
Needed modules
mio
Variables
-
neigh/maxneigh[integer,public/optional/default=3]¶
-
-
neigh/neighcell(*,*,*) [integer,pointer/public]¶
-
-
neigh/neighd(*,*,*) [real,pointer/public]¶
-
-
neigh/nlist(*,*) [integer,pointer/public]¶
-
-
neigh/nlist2(*,*) [integer,pointer/public]¶
-
-
neigh/nneigh(*) [integer,pointer/public]¶
-
-
neigh/nodesndrcv(*,*) [integer,pointer/public]¶
-
-
neigh/nradii(*,*) [real,pointer/public]¶
-
-
neigh/rcvindx(*,*) [integer,pointer/public]¶
-
-
neigh/rcvlist(*) [integer,pointer/public]¶
-
-
neigh/rcvsz[integer,public]¶
-
-
neigh/sndindx(*,*) [integer,pointer/public]¶
-
-
neigh/sndlist(*) [integer,pointer/public]¶
-
-
neigh/sndsz[integer,public]¶
-
Subroutines and functions
-
subroutine
neigh/neighlist()¶
-
subroutine
neigh/neighlistold()¶ - Use
atoms(atomssetcart(),indxdiv(),indxnode()),parallel(xdiv(),ydiv(),procid()),cell(ucell()),math- Call to
-
subroutine
neigh/neighindxcell(isec, secd, insec, ncell, xdiv, ydiv, xsubdiv, ysubdiv, proc, secat)¶ - Parameters
isec [integer,in]
secd (2) [integer,in]
insec (2) [integer,out]
ncell (3) [integer,out]
xdiv [integer,in]
ydiv [integer,in]
xsubdiv [integer,in]
ysubdiv [integer,in]
proc [integer,in]
secat (*) [integer,in]
- Called from
-
subroutine
neigh/fastnnnotsquare(natoms, x, y, z, acc, cutoff2, cutoff2bis, a1, a2, maxnn)¶ - Options
natoms [integer,in,optional/default=len(x)]
- Parameters
x (natoms) [real,in]
y (natoms) [real,in]
z (natoms) [real,in]
acc [real,in]
cutoff2 [real,in]
cutoff2bis [real,in]
a1 (3) [real,in]
a2 (3) [real,in]
maxnn [integer,in]
- Use
atoms(atomssetcart(),frac()),tbpar(tbnn()),cell(ag(),abn(),ucell()),math- Called from
-
subroutine
neigh/fastnnnotsquaresmall(natoms, x, y, z, acc, cutoff2, cutoff2bis, a1, a2, maxnn)¶ - Options
natoms [integer,in,optional/default=len(x)]
- Parameters
x (natoms) [real,in]
y (natoms) [real,in]
z (natoms) [real,in]
acc [real,in]
cutoff2 [real,in]
cutoff2bis [real,in]
a1 (3) [real,in]
a2 (3) [real,in]
maxnn [integer,in]
- Use
atoms(atomssetcart(),frac()),tbpar(tbnn()),cell(ag(),abn(),ucell()),math- Called from
-
subroutine
neigh/fastnnnotsquarebulk(natoms, x, y, z, acc, cutoff2, cutoff2bis, a1, a2, a3, maxnn)¶ - Options
natoms [integer,in,optional/default=len(x)] :: Find the index ix and iy of a binning cell and fill it with all the atoms that are inside there
- Parameters
x (natoms) [real,in]
y (natoms) [real,in]
z (natoms) [real,in]
acc [real,in]
cutoff2 [real,in]
cutoff2bis [real,in]
a1 (3) [real,in]
a2 (3) [real,in]
a3 [real,in]
maxnn [integer,in]
- Use
atoms(atomssetcart(),frac()),tbpar(tbnn()),cell(ag(),abn(),ucell()),math- Called from
-
subroutine
neigh/fastnnnotsquarebulksmall(natoms, x, y, z, acc, cutoff2, cutoff2bis, a1, a2, a3, maxnn)¶ - Options
natoms [integer,in,optional/default=len(x)]
- Parameters
x (natoms) [real,in]
y (natoms) [real,in]
z (natoms) [real,in]
acc [real,in]
cutoff2 [real,in]
cutoff2bis [real,in]
a1 (3) [real,in]
a2 (3) [real,in]
a3 [real,in]
maxnn [integer,in]
- Use
atoms(atomssetcart(),frac()),tbpar(tbnn()),cell(ag(),abn(),ucell()),math- Called from
-
subroutine
neigh/fastnnnotsquarenotrectangle(natoms, x, y, z, acc, cutoff2, cutoff2bis, a1, a2, maxnn)¶ - Options
natoms [integer,in,optional/default=len(x)]
- Parameters
x (natoms) [real,in]
y (natoms) [real,in]
z (natoms) [real,in]
acc [real]
cutoff2 [real]
cutoff2bis [real]
a1 (3) [real]
a2 (3) [real]
maxnn [integer,in]
- Use
atoms(frac(),atomssetcart()),tbpar(tbnn()),cell(ag(),abn(),ucell()),math- Called from
- Call to
-
subroutine
neigh/fastnn(natoms, x, y, z, acc, cutoff, a1, a2, maxnn)¶
parallel.f90¶
Quick access
Needed modules
mio
Variables
-
parallel/ndiv[integer,save/public]¶
-
-
parallel/nth(*) [integer,pointer/public]¶
-
-
parallel/procid[integer,save/public]¶ From 1 to nDiv
-
-
parallel/xdiv[integer,save/public]¶
-
-
parallel/xsubdiv[integer,save/public]¶
-
-
parallel/ydiv[integer,save/public]¶
-
-
parallel/ysubdiv[integer,save/public]¶
-
Subroutines and functions
random.f90¶
Description
http://jblevins.org/log/openmp https://gcc.gnu.org/onlinedocs/gfortran/RANDOM_005fSEED.html
Quick access
- Types
- Variables
1131199299,16163801,287291,299977,301237,362436069,388673,456623,499819,501173,default_seed,ns,prime,rand_t- Routines
Needed modules
mio
Types
Variables
-
random/1131199299[integer,private/parameter]¶
-
-
random/16163801[integer,private/parameter]¶
-
-
random/287291[integer,private/parameter]¶
-
-
random/299977[integer,private/parameter]¶
-
-
random/301237[integer,private/parameter]¶
-
-
random/362436069[integer,private/parameter]¶
-
-
random/388673[integer,private/parameter]¶
-
-
random/456623[integer,private/parameter]¶
-
-
random/499819[integer,private/parameter]¶
-
-
random/501173[integer,private/parameter]¶
-
-
random/default_seed(4) [integer,private/parameter/optional/default=[521288629]¶
-
-
random/ns[integer,private/parameter/optional/default=4]¶
-
-
random/prime(8) [integer,private/parameter/optional/default=[99991]¶
-
-
random/rand_t[public]¶
-
Subroutines and functions
scf.f90¶
Quick access
Needed modules
mio
Variables
-
scf/charge(*,*) [real,pointer/public]¶
-
-
scf/dwork(*) [real,private/pointer/optional/default=>null()]¶
-
-
scf/lwork[integer,private]¶
-
-
scf/zch[real,public/parameter/optional/default=1.0_dp]¶
-
-
scf/zwork(*) [complex,private/pointer/optional/default=>null()]¶
-
Subroutines and functions
-
subroutine
scf/scfgetcharge()¶
-
subroutine
scf/scfham0(n, h, e, k, cell, h0, maxn, hopp, nlist, nneigh, neighcell)¶ - Options
n [integer,in,optional/default=len(h0)]
- Parameters
h (n,n) [complex,out]
e (n) [real,out]
k (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
- Use
- Called from
-
subroutine
scf/scfham(n, h, e, charge, ns, k, cell, h0, maxn, hopp, nlist, nneigh, neighcell, species)¶ - Parameters
n [integer,in,]
h (n,n) [complex,out]
e (n) [real,out]
charge (2,n) [real,in]
ns [integer,in]
k (3) [real,in]
cell (3,3) [real,in]
h0 (n) [real,in]
maxn [integer,in,]
hopp (maxn,n) [complex,in]
nlist (maxn,n) [integer,in]
nneigh (n) [integer,in]
neighcell (3,maxn,n) [integer,in]
species (n) [integer,in]
- Use
- Called from
-
subroutine
scf/scfinit(n)¶ - Parameters
n [integer,in]
- Called from
tbpar.f90¶
Quick access
Needed modules
mio
Variables
-
tbpar/e0_b[real,save/public]¶
-
-
tbpar/e0_c[real,save/public]¶
-
-
tbpar/e0_c1[real,save/public]¶
-
-
tbpar/e0_c2[real,save/public]¶
-
-
tbpar/e0_n[real,save/public]¶ On-site energy for carbon, boron and nitrogen
-
-
tbpar/g0[real,save/public]¶ Hopping parameter
-
-
tbpar/gintlay[real,save/public]¶
-
-
tbpar/gn(*,*,*) [real,pointer/public]¶
-
-
tbpar/tbnn[integer,save/public]¶
-
-
tbpar/u(4) [real,save/public]¶
-
Subroutines and functions
-
subroutine
tbpar/tbinit()¶ - Use
- Called from
version.f90¶
Quick access
- Variables
- Routines
Variables
-
version/update[character,private/parameter/optional/default=”kubo_update”]¶
-
-
version/ver[character,private/parameter/optional/default=”kubo_version”]¶
-
Subroutines and functions
-
function
version/versionget()¶ - Return
res [character]
-
function
version/versionupdate()¶ - Return
res [character]